10.2. Introduction to Networkx¶
Note
Most of the points covered here are covered in
an ipython notebook (.ipynb).
Help with the homework assigned for this module is also available there.
In this section we present a very brief introduction
to networkx, one of the more widely used Python
tools for network analysis.
For the code below to work you must have installed
packages named networkx and
(for graphviz) pydot. These
are part of the standard Canopy distribution.
Some additional networkx help:
For a tutorial on using
networkx, look here. Detailed documentation ofnetworkxand many resources and tutorials are available at the Networkx website. But use the Enthought package manager rather than the website for installation.This Rice University Tutorial. is very stripped down and very well done.
10.2.1. Basics¶
We will import as follows:
>>> import networkx as nx
Networkx provides some graphs as examples. One of these happens to be Zachary’s karate club. That can be read in as follows:
>>> kn=nx.karate_club_graph()
In general, you will be reading in graphs that you construct yourself
and save in a file in some standard format or that are available as
course data. An alternative way of constructing Zachary’s graph is to download Zachary's karate club network
and save this in the directory you start Python in. GML format is a standard
graph format networkx knows about, so that file can be read into Python
as follows:
>>> kn = nx.read_gml('karate.gml')
<networkx.classes.graph.Graph object at 0x1014b7f90>
You will often find it useful to do:
>>> filename = 'yourfile.gml'
>>> G = nx.read_gml(filename,relabel=True)
The relabel=True flag builds a graph that
uses names (actually, whatever occurs as value of the label
attribute) as node identifiers.
For example, when reading in the Les Miserables graph
or the Anna Karenina graph, this will give you characters
identified by their two character IDs.
networkx also provides a number of methods
that compute statistics of your graph, many of which
we will discuss below. To get the number of nodes in your graph,
for example, do:
>>> num_nodes = G.number_of_nodes()
>>> print 'number of nodes: ' + str(num_nodes)
10.2.2. Graph generators in networkx1¶
Networkx provides a number of functions that generate famous
graphs. We have already seen one example. Zachary’s
karate club graph can be generated with a simple
function call:
>>> kn=nx.karate_club_graph()
To create a Watts-Strogatz (WS) small world graph, do:
>>> G = nx.watts_strogatz_graph(24,4,.3)
G is a WS-graph with 24 nodes, each of which is linked to its four nearest neighbors in the original circular layout. The probability of rewiring is set to .3. Now draw the result.
>>> nx.draw_circular(G)
>>> plt.show()
To create an erdos-renyi graph with 80 nodes, and edge connection probability .1, do:
>>> nx.gnp_random_graph(80,.1)
For more examples of graphs that can be generated, see the Networkx docs.
10.2.3. A simple example¶
As our first example, we will reproduce the result discussed in the visualization section, in which we visualize Zachary’s karate club in a way which that shows the factionalized nature of the social relationships.
First, download Zachary's karate club network
and save it in your Python directory, if you haven’t already. Then
read it into Python:
>>> kn = nx.read_gml('karate.gml')
You can now draw the graph:
>>> import matplotlib.pyplot as plt
>>> nx.draw_circular(kn)
If you are not working in Ipython with pylab active, you will
need to execute the show command to actually display the graph:
>>> plt.show()
You may want to make some small adjustments to the display in the display window. You can then save the figure in an image file in various formats. Here we choose “png”, a standard webbrowser friendly format:
>>> plt.savefig('networkx_circular_karate.png')
10.2.4. Graph analysis¶
networkx has a standard dictionary-based
format for representing graph analysis computations
that are based on properties of nodes.
We will illustrate this with the example of
betweenness_centrality.
The problem of centrality and the various ways
of defining it was discussed in Section Social Networks.
As noted there, key facts about the karate graph can be revealed with
a measure of graph centrality called betweenness centrality.
networkx offers an implementation, along with a host of
other graph analysis tools:
>>> betweenness = nx.betweenness_centrality(kn)
Like the other centrality
measures discussed below,
betweenness_centrality returns
a dictionary associating each node in the graph with
a betweenness score. Higher is better. That is, higher means
that node is evaluated as occupying a more central position in
the graph:
1>>> betweenness
2{1: 0.43763528138528146, 2: 0.053936688311688304, 3: 0.14365680615680618,
3 4: 0.011909271284271283, 5: 0.0006313131313131313, 6: 0.02998737373737374,
4 7: 0.029987373737373736, 8: 0.0, 9: 0.05592682780182781, 10: 0.0008477633477633478,
5 ...
6}
Computing centrality measures is often followed by sorting the results to find the nodes with highest centrality. Here’s an example of how to do that. Dictionaries have no concept of order, so to sort their contents you must first turn them into lists, and then sort the resulting list:
1>>> b_il = betweenness.items()
2>>> b_il.sort(key=lambda x:x[1],reverse=True)
3>>> b_il
4[(1, 0.43763528138528146), (34, 0.30407497594997596), (33, 0.14524711399711399),
5 (3, 0.14365680615680618), (32, 0.13827561327561325), (9, 0.05592682780182781),
6 (2, 0.053936688311688304), (14, 0.04586339586339586), (20, 0.03247504810004811),
7 ....
8]
So we’ve discovered that node 1 and node 34 have the highest betweenness scores, and might therefore be thought to be the most central people in this social organization, in accord with what is suggested by the display produced by the Fruchterman-Reingold algorithm.
We can compare the results obtained from betweenness_centrality with
those from degree_centrality:
>>> deg = nx.degree_centrality(kn)
The results in this case aren’t very different from those of betweenness centrality, so we omit them here.
Other analysis tools implemented in networkx.
edge_betweenness(G):Illustrated below in the the Girvan-Newman example.eigenvector_centrality(G): (also eigenvector_centrality_numpy). Explaining this concept of centrality is beyond the scope of this course. It requires computing the eigenvectors of the adjacency matrix of the graph, and is closely related to pagerank score used by Google to rank the centrality of websites on the Internet.nx.closeness_centrality(G). Closeness centrality of a node u is the reciprocal of the sum of the shortest path distances from u to all n-1 other nodes. Since the sum of distances depends on the number of nodes in the graph, closeness is normalized by the sum of minimum possible distances n-1. So if node n is a neighbor of all n-1 other nodes in the graph, closeness_centrality is 1.current_flow_betweenness_centrality. This kind of centrality was first discussed in [NEWMAN2003] . Newman calls it “random walk centrality”, also known as “current flow centrality”. The current flow centrality of a node n can be defined as the probability of passing through n on a random walk starting at some node s and ending at some other node t (neither equal to n), Newman argues that betweenness for some social networks should not be computed just as a function of shortest paths, but of all paths, assigning the shortest the greatest weight. This is because messages in in such networks may get to where they’re going not by the shortest path, but by a random path, such as that of sexual disease through a sexual contact network.nx.clustering coefficient(G): clustering(G, nbunch=None, with_labels=False, weights=False) Clustering coefficient for each node in nbunch.nx.average_clustering(G): Average clustering coefficient for a graph. Note: this is a space saving function ; It might be faster to use clustering to get a list and then take average.nx.triangles(G, nbunch=None, with_labels=False): Returns number of triangles for nbunch of nodes as a dictionary. If nbunch is None, then return triangles for every node. If with_labels is True, return a dict keyed by node. Note: Each triangle is counted three times: once at each vertex. Note if G is the graph, and t is thenumber triangles of a node n, and d is its degree, then :math: nx.clustering_coefficient(G,n) = frac{t}{d(d-1)}nx.connected_component_subgraphs(G): Used in the Girvan-Newman implementation below. This returns a list of the fully connected components of graph G. If the graph is fully connected it returns a list containing the graphnx.number_connected_components(G): This just returns the length of the list returned bynx.connected_component_subgraphs(G).
10.2.5. Implementing Girvan-Newman¶
Here is a simple python implementation of the Girvan-Newman graph-partition
method using networkx. The source code is
here.
The Girvan-Newman algorithm is divisive. It tries to find an optimal
way of cutting the graph into two pieces, and then it does the same on
the pieces. Hence the girvan_newman function is recursive.
Each time it finds a way to split a graph into two optimal
pieces, it calls itself on the pieces. It terminates when it is given
a graph with only one node. It returns a list of all the components
it has found:
1import networkx as nx
2
3def girvan_newman (G):
4
5 if len(G.nodes()) == 1:
6 return [G.nodes()]
7
8 def find_best_edge(G0):
9 """
10 Networkx implementation of edge_betweenness
11 returns a dictionary. Make this into a list,
12 sort it and return the edge with highest betweenness.
13 """
14 eb = nx.edge_betweenness_centrality(G0)
15 eb_il = eb.items()
16 eb_il.sort(key=lambda x: x[1], reverse=True)
17 return eb_il[0][0]
18
19 components = nx.connected_component_subgraphs(G)
20
21 while len(components) == 1:
22 G.remove_edge(*find_best_edge(G))
23 components = nx.connected_component_subgraphs(G)
24
25 result = [c.nodes() for c in components]
26
27 for c in components:
28 result.extend(girvan_newman(c))
29
30 return result
Note the use of a
while loop in line 21. This keeps removing
the remaining edge with highest betweenness. Each time an edge is removed
we recompute the edge with highest betweenness as well as the
components in the graph. When we get two components,
the condition for re-entering the while loop fails,
and we exit the loop, having
succeeded in splitting the graph. For densely connected
graphs, many edges may have to be removed to split
the graph.
10.2.6. Using weighted links¶
We read in a weighted version of the karate graph, which has Zachary’s original
weights for the intensity of the relationship (this was measured by
how many extra-club activities the two club members shared):
import networkx as nx
kn = nx.read_gml('weighted_karate.gml')
Nex we sort the edge list by weight
all_edges = sorted(kn.edges(), key= lambda x: kn.get_edge_data(*x)[0]['value'], reverse=True)
Now for each edge we print the edge and its weight:
1from collections import Counter
2all_node_ctr = Counter()
3for (u,v) in all_edges:
4 for x in (u,v):
5 all_node_ctr[x] += 1
6 print '%-4s %-4s %.3f' % (u,v,weight)
126 32 7.000
226 32 7.000
32 3 6.000
42 3 6.000
51 3 5.000
61 3 5.000
7.....
Next we sort by degree:
1all_nodes_il = sorted(all_node_ctr.items(), key = lambda x:x[1], reverse=True)
2for (char, ct) in all_nodes_il:
3 print '%4s %.3f' % (char, ct)
4
5top_chars_degree_centrality = all_nodes_il[:12]
6print
7for (ch, ct) in top_chars_degree_centrality:
8 print '%4s %.3f' % (ch, ct, )
Now we draw. We’ll use a force-spring layout, and we’ll try increasing spring tension for high weighted edges
1new_labels_kn = dict([(char,char) for (char,score) in top_chars_degree_centrality])
2pos = nx.spring_layout(kn,scale=1.0,weight='value') #
3nx.draw_networkx(kn,pos, with_labels=False,node_size=200,node_color="lightgray") #Default node size=300
4nx.draw_networkx_labels(kn,pos,new_labels_kn,font_size=14,font_color='blue')
5nx.draw_networkx_nodes(kn,pos,new_labels_kn,node_color='red',node_size=400,label_pos=1.0,node_shape='D',font_size=16)
10.2.7. Coloring factions¶
The networkx version of the karate graph has
nodes annotated with faction membership information.
Let’s use that to color our factions.
The following are the steps taken to produce the colored FG layout figure in the lecture notes. Some of the commented out lines show experiments you could try.
Start with faction1_color as the default node color We make a list of L tokens of the faction1_color string, where L is the length of kn.nodes()
1#nx.draw_spring(kn)
2#plt.savefig('networkx_spring_karate.png')
3
4import math
5
6#(faction1_col, faction2_col) = ('lightgray','salmon')
7(faction1_col, faction2_col) = ('yellow','lightgreen')
8
9node_color = [faction1_col] * len(kn.nodes())
10# Now using the faction membership abnnotations from Zachary's data, change the known faction2 nodes to faction2_color.
11
12node_dict = dict(kn.nodes(data=True))
13for n in kn.nodes():
14 if node_dict[n]['club'] == 'Officer':
15 node_color[n] = faction2_col
16print node_dict
17print node_color
{0: {'club': 'Mr. Hi'}, 1: {'club': 'Mr. Hi'}, 2: {'club': 'Mr. Hi'}, 3: {'club': 'Mr. Hi'}, 4: {'club': 'Mr. Hi'}, 5: {'club': 'Mr. Hi'}, 6: {'club': 'Mr. Hi'}, 7: {'club': 'Mr. Hi'}, 8: {'club': 'Mr. Hi'}, 9: {'club': 'Officer'}, 10: {'club': 'Mr. Hi'}, 11: {'club': 'Mr. Hi'}, 12: {'club': 'Mr. Hi'}, 13: {'club': 'Mr. Hi'}, 14: {'club': 'Officer'}, 15: {'club': 'Officer'}, 16: {'club': 'Mr. Hi'}, 17: {'club': 'Mr. Hi'}, 18: {'club': 'Officer'}, 19: {'club': 'Mr. Hi'}, 20: {'club': 'Officer'}, 21: {'club': 'Mr. Hi'}, 22: {'club': 'Officer'}, 23: {'club': 'Officer'}, 24: {'club': 'Officer'}, 25: {'club': 'Officer'}, 26: {'club': 'Officer'}, 27: {'club': 'Officer'}, 28: {'club': 'Officer'}, 29: {'club': 'Officer'}, 30: {'club': 'Officer'}, 31: {'club': 'Officer'}, 32: {'club': 'Officer'}, 33: {'club': 'Officer'}}
['yellow', 'yellow', 'yellow', 'yellow', 'yellow', 'yellow', 'yellow', 'yellow', 'yellow', 'lightgreen', 'yellow', 'yellow', 'yellow', 'yellow', 'lightgreen', 'lightgreen', 'yellow', 'yellow', 'lightgreen', 'yellow', 'lightgreen', 'yellow', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen', 'lightgreen']
Next lay out the network:
pos = nx.spring_layout(kn,scale=1.0)
Now put in the labels, using a label->new_label mapping. We’ll change the labels to agree with
Zachary’s original indexing, so 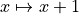 .
.
new_labels = dict((x,x + 1) for x in kn.nodes())
nx.draw_networkx_labels(kn,pos,new_labels,font_size=14,font_color='black')
We’ll highlight the two leader nodes, by drawing slightly larger black circles round them. Notice how the edges and nodes have to be drawn separately now.
nx.draw_networkx_nodes(kn,pos,{0:0,33:33},node_color=['black','black'],node_size=800)
# Now draw all the nodes, including leaders, using faction color scheme.
nx.draw_networkx_nodes(kn,pos,new_labels,node_color=node_color,node_size=500)
nx.draw_networkx_edges(kn,pos)
10.2.8. Notes on color¶
For your choices with matplotlib colors, look here in the matplotlib docs. Or you can use direct html color specs replacing line ?? above with something like this:
>>> (faction1_col, faction2_col) = ('#00CCFF','#FF00FF')
This following table shows the HTML color scheme:
10.2.9. Using the graphviz interface¶
Here is another way to access tools outside of
Python for graph drawing, in particular
a popular graph drawing program named graphviz.
Via the module pydot networkx provides an interface
to graphviz. This
is loaded automatically whenever graphviz -relevant functions
are called. For example:
>>> nx.write_dot(kn,'networkx_circular_karate.dot')
This writes a “.dot” file. This is a layout neutral representation
of the graph. Now graphviz can be called with various layout
functions to create an image file from the the graph
representation in networkx_circular_karate.dot.
The most interesting result for the karate graph was via the graphviz circo function:
circo -Teps -o networkx_circular_karate_via_dot.eps networkx_circular_karate.dot
The result looks like this.
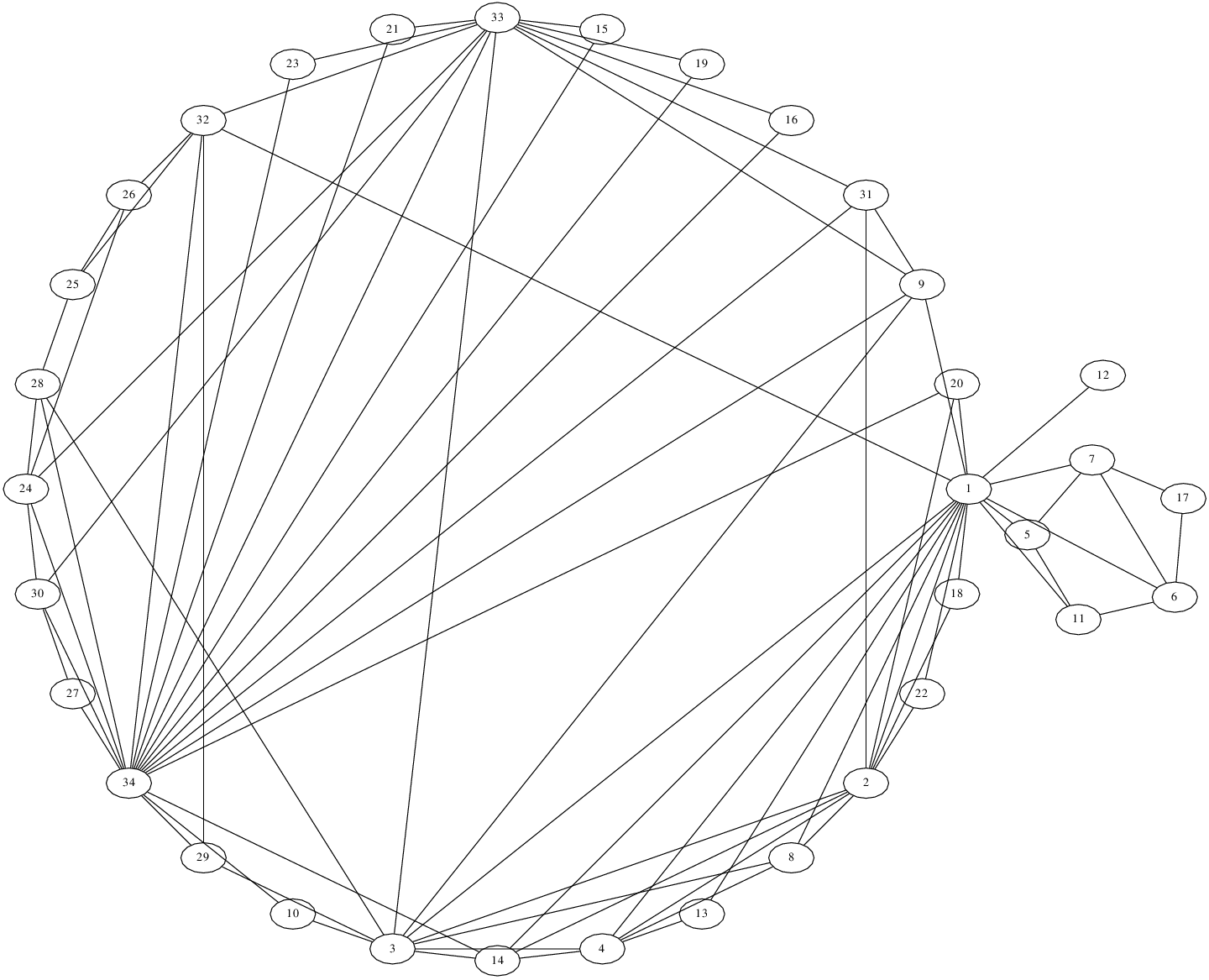
networkx also offers the option of calling
graphviz layout programs directly and plotting them using
matplotlib. For example, graphviz ‘s version of
a Fruchterman-Reingold layout can be computed
as follows:
>>> from networkx import graphviz_layout
>>> layout=nx.graphviz_layout
>>> pos = layout(pb,prog='sfdp')
Then networkx ‘s interface to plotting facilities can be called.
